
Features
Agronomy
Other Crops
On the threshold of transformation
Genomics, the study of an organism’s complete DNA sequence, is poised to transform the life sciences, including agriculture, forestry, human health and the environment. For crops, genomics could result in major leaps in varietal improvement.
September 17, 2010 By Carolyn King
|
|
| Canola’s importance as an oilseed crop for food, feed and bio-products has spurred canola genomic research by many public and private agencies. Photo courtesy of Genome Prairie.
|
Genomics, the study of an organism’s complete DNA sequence, is poised to transform the life sciences, including agriculture, forestry, human health and the environment. For crops, genomics could result in major leaps in varietal improvement. And it could open new doors into value-added markets such as nutraceutical, pharmaceutical and industrial uses for crops. “In the 1980s and early ’90s, we had learned how to identify and manipulate single genes and to transfer a single gene into a crop. For example, insect-resistant corn and herbicide-tolerant canola involved insertion of single genes,” says Dr. Wilf Keller. Keller has years of experience in cell genetics, plant biotechnology and crop genomics, and is currently president and chief executive officer of Genome Prairie (see “Genome Canada and its Prairie centres,” below). “However, many crop traits, including drought tolerance, cold tolerance and seed size, are far more complex and require the management and manipulation of many different genes.
Without full knowledge of that system, it would be very hard to know how to direct a plant improvement or plant breeding strategy. The genomics approach gives an overview of the whole system.”
He draws an analogy: “If you are trying to improve the way a car works, the single gene approach is like putting in a better spark plug to get better performance. In the genomics approach where you map the whole genome, you stand back and look at the whole car; the size of the wheels, the types of tires, the size of the motor, the aerodynamic shape of the car, and so on. All of these things are combined in your analysis to design a vehicle that performs much better.”
Similarly, considering the whole genome of an organism, whether it is a plant, an animal or a human, could lead to significant advances. Keller says, “That’s why I’m excited about the future of the life sciences.”
 |
|
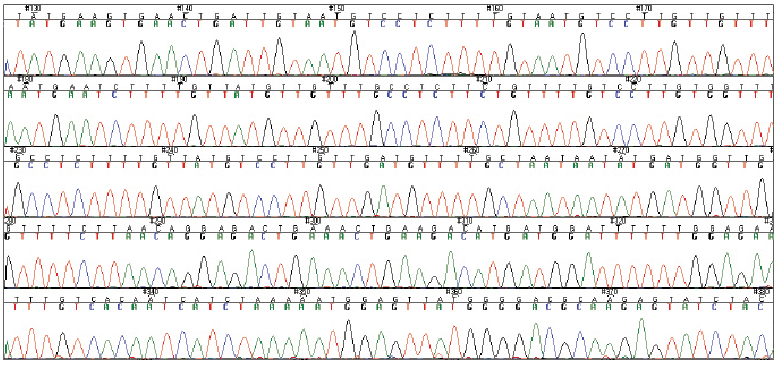
| This is a “sequence trace file” that shows the sequence of a piece of Brassica DNA. The building blocks of DNA are four nucleotides: adenine (A), guanine (G), thymine (T) and cytosine (C). An organism’s DNA has many of each of the four types. DNA sequencing is the process of determining the order of the nucleotides in the DNA. In the sequence trace file shown here, each of the nucleotides is shown as a different coloured peak, with the name of the nucleotide (A, G, T or C) shown above the peak.Photo courtesy of Isobel Parkin, AAFC. |
Mapping and managing life’s building blocks
A central aspect of genomics is identifying what specific genetic material is where along an organism’s DNA. “DNA sequencing” is the process of determining the exact order of all the chemical building blocks that make up an organism’s entire DNA. “Genome mapping” identifies landmarks, specific DNA sequences known as “genetic markers,” along the DNA.
Keller explains, “Knowing the complete DNA sequence of an organism provides an understanding of how all the different genetic components interact with each other, and how they are involved in defining the properties of the organism. “Let’s use a crop as an example. Genetic information is required for a crop to grow from a seed through to its development, maturity and producing another set of seeds. A lot of genetic elements or genes are involved during the life cycle of that crop; from the composition of the oil in the seed, to the resistance to diseases, to the ability to take up fertilizer out of the soil. By mapping the genome, you get an understanding of how all of these genes are ordered in sequence with each other, how many are ultimately required to exhibit a given trait.”
|
|
| INSET: Genomics, the study of an organism’s complete DNA sequence, can be used along with conventional breeding or genetic engineering to develop significant improvements in crop traits. Photo courtesy of Genome Prairie. |
|
 |
|
| Dr. Habibur Rahman, a canola breeder at the University of Alberta and one of the Designing Oilseeds for Tomorrow’s Markets (DOTM) researchers, is using breeding strategies to gain insight into factors affecting canola seed coat thickness, which affects fibre content in canola meal. Photo courtesy of University of Alberta. | |
 |
|
| DOTM personnel with Randall Weselake’s group at the University of Alberta (left to right): plant molecular biologists Gavin Chen and Wei Deng, computational biologist Fred Peng, and program manager Disa Brownfield-Walker. Photo courtesy of Dr. Randall J. Weselake, University of Alberta. | |
 |
|
| Flax seeds, from Dr. Gordon Rowland’s program, represent diverse flax collections and genotypes. Photo courtesy of Prakash Venglat and Raju Datla. | |
 |
|
| Genomics could help flax breeders develop varieties tailored to specific market needs. Photo courtesy of Genome Prairie.
|
Once a crop’s genome is mapped or sequenced, this genomic information can be used along with conventional breeding or genetic engineering applications to develop varieties that meet the needs of growers, processors and consumers.
For example, researchers could determine which genes are associated with a desired trait and investigate how those different genes work together to express that trait. They could develop genetic markers to screen new breeding lines for the desired genes. They could use that information to produce improved crop lines more quickly or to enhance a trait by a much larger increment. They could identify very small variations in the DNA between individual plants, and explore how those small variations affect certain traits. They might discover novel traits in individual plants that could allow development of varieties with new strengths and new uses.
As well, genomic techniques are getting quicker and less expensive. For instance, in less than a decade, DNA sequencing costs have dropped from hundreds of millions of dollars to thousands of dollars. So, advances through genomics will likely come increasingly faster as scientists sequence the DNA of more and more organisms, as the library of genomic information grows, and as genomic tools are continually improved.
Keller says improvements in crops, livestock and human health through genomics could be “transformational” in our society. For crops, he envisions significant improvements in complex traits like yield, disease tolerance and drought tolerance within the next 10 to 15 years, as well as many other potentially far-reaching advances.
“I see a major transformation coming in terms of significant improvements in the quantity and the quality of our food; we need to feed nine billion people by 2040. By producing food with components that reduce the incidence of diabetes and cardiovascular diseases, we reduce health care costs through prevention rather than treatment. I see the opportunity for using plants to produce a wide range of environmentally friendly industrial products, including fibres, polymers, nylon and bio-plastics, and to produce drugs and pharmaceutical products. Plants might even be designed to remediate or detoxify contaminated soils where there have been serious spills or pollution.”
Will all this potential opportunity become a reality for Canadian crop producers? Keller cautions, “Every discovery, if you want to turn it into a product, a treatment or a drug, or a new food product on your plate, has to involve an interface with companies and industries to pick up those ideas and to move forward with them. I think Canada needs to be very committed to looking at that pipeline going from discovery right through the various stages to product. I think we need to, as a society, figure out where we make the investments, and where we streamline our regulatory procedures so we can get these things through so that our citizens benefit from them, as opposed to losing the technology to other countries and then we import it back in.”
Genome Canada and its Prairie centres
Genome Canada is a not-for-profit agency implementing a national genomics strategy to benefit all Canadians. Since 2000, it has been supporting genomics research in agriculture, fisheries, forestry, human health, the environment and social issues, as well as genomics technology development. It has six regional centres to support and manage its projects, including Genome Prairie with responsibilities for projects in Manitoba and Saskatchewan, and Genome Alberta for projects in Alberta.
Genome Canada-supported research on the Prairies includes projects on plants, like canola and flax, and on other such topics as cattle genome sequencing, improving diagnosis and management of human diseases, and greener production of hydrocarbon energy.
Sequencing as a wellspring for advancing canola
Canola’s importance as an oilseed crop used for food, feed, biofuel and other bio-products has spurred efforts by many agencies to sequence its genome. In October 2009, Bayer CropScience announced it had sequenced canola in collaboration with several partners; however that sequence is not publicly available. So other researchers are still working to sequence canola. One such project is the Canadian Canola Sequencing Initiative (CanSeq).
CanSeq is a partnership of Agriculture and Agri-Food Canada (AAFC), the Plant Biotechnology Institute of the National Research Council (NRC), Genome Alberta and several private companies. The project’s co-leaders are Dr. Isobel Parkin from AAFC and Dr. Andrew Sharpe from the NRC.
Canola, or Brassica napus, has a large, complicated genome with about 60,000 genes. Parkin says Brassica napus evolved from “the fusion of a turnip and a cabbage,” so it is effectively two genomes stuck together. “The turnip part of the genome is represented by Brassica rapa, and the cabbage part is represented by Brassica oleracea. These modern-day relatives are the best representations we have of the ancestors of B. napus. If you put B. rapa and B. oleracea together, you are effectively looking at canola.”
CanSeq, working with researchers in other countries, is sequencing Brassica rapa and B. oleracea to provide a foundation, and then sequencing canola. Parkin expects they will likely release the B. rapa genome to the public domain in fall 2010, B. oleracea in spring 2011, and canola by the end of 2011.
Along with sequencing these genomes, CanSeq is “re-sequencing” B. rapa and B. napus lines. Parkin explains, “When you sequence your first genome, you try to do the best job you can with the resources you have to get the best foundation. Then with re-sequencing you take a much quicker look at the genome of a number of different genotypes that vary in phenotype and try to associate any variation at the sequence level with variation in phenotype.”
A genotype is an individual organism’s full genetic information; for example, if one canola plant has differences in its DNA compared to another canola plant, then they are different genotypes. A “phenotype” is an individual organism’s observable traits, like a plant’s seed colour or oil content. For example, a tall canola plant and a short one would be two different phenotypes. Phenotype is influenced by the organism’s genetics and its environment.
In fact, if someone wanted to figure out which genes are associated with canola seed size (an observable trait), it would require re-sequencing many lines with large seeds and many lines with small seeds and then comparing their DNA sequences to see where they differ.
Parkin notes, “As soon as we’ve released the initial genome for canola, other people will also be able to look at their own lines of interest and start capturing the information in those lines and find out what it means. Hopefully other researchers will also contribute to data in the public domain, so that gradually there will be a database of sequences of different genotypes that people can access.”
Sequencing of the canola genome opens the way to using various genomic tools to speed the development of improved varieties. Parkin gives some examples: “When you’re trying to identify genes within a genome where the sequence is already available, then you don’t have to do upstream work to isolate the gene. You just search in a database to find your gene. And, as we start re-sequencing new genotypes, we will find associations between sequence variation and particular phenotypes of interest. Then we could facilitate the breeding process, using markers to rapidly identify the traits of interest and combine them in single lines. We could make lines that perform better in the field and do it more rapidly.”
She adds, “In canola it’s probably going to have value in hybrid development because it is often difficult to predict which lines are going to give you better hybrid potential. But if you have more information at the sequence level and information about which combinations work to give you good hybrids, then you could hopefully predict which lines to combine to make better hybrids.”
Functional genomics for improving canola meal
While CanSeq is using DNA sequencing to decode canola’s genome, another project is using “functional genomics” to understand and enhance canola seed traits for the livestock and aquaculture feed markets. “DNA sequencing technology makes all the information in an organism’s genetic blueprint available to you. The information in that blueprint is converted into various entities in the cell, like enzymes and proteins, which have various functions in the cell. The ability to see how the information from the blueprint flows into cell functions is called ‘functional genomics.’ It’s somewhat like having a blueprint for a house, and determining the flow of activity, all the workers doing their jobs, to make that blueprint into a functioning house,” explains Dr. Randall Weselake from the University of Alberta.
Dr. Disa Brownfield-Walker, also with the University of Alberta, adds, “Essentially, you are trying to get from the blueprint to the house, and so you do a series of snapshots, at different time points and with different genotypes, to get a better idea of the whole program of events.”
That timed series of snapshots enables researchers to see which genes are involved at each step as a canola seed develops into a mature seed.
Dr. Weselake and Dr. Gopalan Selvaraj, from the National Research Council (NRC)’s Plant Biotechnology Institute are co-leading the project, called Designing Oilseeds for Tomorrow’s Markets (DOTM). Brownfield-Walker is DOTM’s project manager. The research team involves Canadian and German researchers, and project funding is from Canadian and German agencies, including Genome Canada/Genome Alberta and the provinces of Alberta and Saskatchewan.
DOTM’s aim is to enhance the nutritional value of canola meal by reducing four anti-nutritional factors, and increasing the carotenoids in canola seed. Canola meal is what remains after the oil is extracted from canola seed. The meal is previously used livestock feed, but the researchers want to make it even better. Weselake outlines some of the issues related to the four anti-nutritional factors: phytate, sinapine, fibre and glucosinolates. Phytate can bind certain nutrients in the meal, preventing them from entering the animal’s nutritional stream. As well, monogastrics, such as pigs, have trouble digesting phytate (a form of phosphorus), so more phosphorus ends up in their manure and could contribute to pollution. Sinapine imparts a bitter flavour to canola meal, and for some types of poultry, a sinapine-rich diet can lead to smelly eggs. High fibre levels can reduce an animal’s ability to absorb energy from the meal. Glucosinolates can be converted into anti-nutritional compounds in the meal.
Carotenoids are important for animal and human health, and offer an additional potential benefit in aquafeed. Weselake says, “Carotenoids result in orange-coloured canola oil. The idea is this oil could be added back to the meal, and if you feed it to a fish, like a salmon, the carotene will help turn the salmon’s flesh to a deeper orange, adding visual appeal for consumers and enhancing the nutritive value.”
By using functional genomics to understand the cell processes affecting these five factors, the researchers have made important progress since DOTM started in 2006. They have substantially increased the carotenoids, and significantly reduced sinapine, glucosinolates and phytate, thus setting the stage for developing canola germplasm; breeding material that can be used to create improved varieties.
Brownfield-Walker notes, “Several collaborators in our project are involved in breeding aspects. The idea is to use their information and our information and, through breeding, modify the canola. So it’s a complementary approach.”
Other Canadian canola genomics research is advancing traits like crop yield, vigour and oil content. Weselake sees genomics as a key element in improving Canada’s canola varieties. “We call canola the “Cinderella” crop. It was developed in Canada, and it’s our major oilseed crop. We contribute up to 20 percent of the world’s canola and rapeseed. I think it’s very important for Canada to maintain a competitive edge, and continue to improve the oil and the meal to improve the overall seed even further and diversify for different uses, or else we are going to lose ground.”
Enhancing value-added opportunities for flax through genomics
Flax is a truly versatile Prairie crop. Its seed and oil are nutritious food ingredients, its oil has industrial uses in linoleum and other products, and the tough, durable fibre from its straw has applications in manufacturing. To make the most of all that flax can offer, a project is investigating flax at the genomic level.
Called Total Utilization Flax Genomics (TUFGEN), this four-year project started in fall 2009. Dr. Gordon Rowland of the University of Saskatchewan and Dr. Sylvie Cloutier of Agriculture and Agri-Food Canada (AAFC)’s Research Centre in Winnipeg are co-leading TUFGEN’s multi-agency research team. The project is funded by Genome Canada/Genome Prairie and many other partners, including federal and provincial agencies and flax grower associations.
The researchers are sequencing the flax genome and mapping the position and relationships of the genes related to the crop’s oil characteristics, such as its fat composition and protein content, and its fibre characteristics, such as yield, extractability and quality.
And they are developing genomic tools to help flax breeders develop new varieties faster and more efficiently. For example, they are developing tools to quickly search for and isolate genes of interest. That is very helpful because a crop plant’s DNA contains thousands of genes, and those genes are only a small percentage of the plant’s total DNA.
Rowland explains, “Take, for example, oleic acid, one of the fatty acids in flax seed. If we can tag the portion of the DNA related to oleic acid, we then could develop a DNA marker to follow that particular trait through the various generations that arise out of crossing various flax plants. So, rather than having to look at the phenotype of the eventual plant, which in this example would be the actual oleic acid, we can follow that trait at the molecular level and follow it much earlier, and know which plants have a better probability of having a high oleic content. It would save a lot of time in the field or in multiple generations in the growth chamber for finding out whether a plant actually carries the desired trait.”
Sequencing the flax genome could also be a springboard for other scientific advances. “One reason DNA sequencing of flax is important is that flax is not closely related to any other crop plant that we know about. And from the way that it arranges itself genetically, we think is quite different from other plant species. So its genome will be of great interest to a lot of people, and may explain some of the things we haven’t understood about flax in the past,” says Rowland.
“For example, there’s a phenomenon in some fibre flax varieties; when they are grown under certain conditions, the flax will actually end up changing its appearance in the next generation, and these changes are then inherited and carry on generation after generation after that. We never could understand how this happens. The more information we get about the plant at the DNA level, I think the more we’ll come to understand how these changes actually occur.”
Rowland believes the potential of genomics to pinpoint specific traits, discover novel traits and make breeding more efficient could help flax breeders develop varieties tailored to specific market needs. “Right now we are producing flax really for just the oil market. I think a number of different markets will come out of this work, for both oil and fibre. For instance we could have flax types that are specific for human health or animal health. I think there will even be submarkets, with types that are high in linolenic acid and types that are high in a compound called lignan, for human health and nutraceutical uses. “We’ll still have the traditional industrial uses for the oil, like linseed oil, paint and linoleum, but there may be other industrial applications for other mixtures of fatty acids produced in flax oil. On the straw side, there could be flax types with high fibre content, perhaps grown for the seed with the fibre as a byproduct or grown specifically for fibre markets.”
Canada is currently the world’s largest flax producer, and Rowland would like to see Canada maintain that strength. He says, “Flax is really important in Canada as a crop but not very important in most other areas of the world. So I think we need to learn as much about flax genomics as we can for this country.”