Features
Agronomy
Diseases
New strains of clubroot identified
Clubroot continues to be a disease risk to canola crops across Alberta, with more than 2,700 clubroot-infested fields now confirmed, along with a few isolated cases in Saskatchewan and Manitoba. Although good clubroot resistant (CR) varieties have been available for about 10 years, there are an increasing number of fields in Alberta where new strains of the pathogen Plasmodiophora brassicae have overcome the resistance.
October 17, 2018 By Donna Fleury
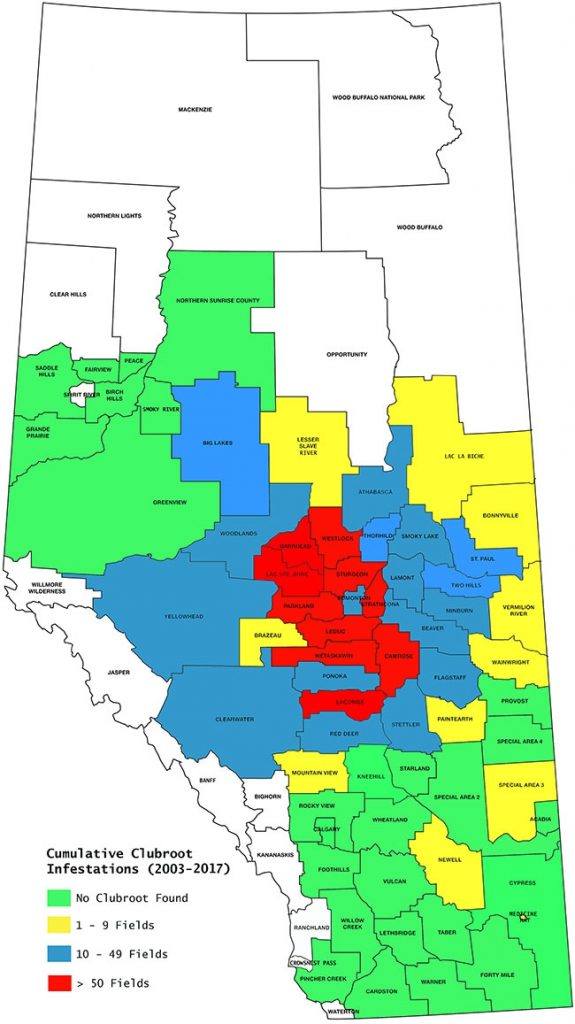 Alberta Clubroot Distribution Map New strains of the pathogen have overcome clubroot resistance.
Alberta Clubroot Distribution Map New strains of the pathogen have overcome clubroot resistance.Particularly in fields under short or intensive canola rotations, inoculum levels have been building up. Increasing inoculum levels allow the pathogen to evolve and select for new pathotypes or strains that appear to be highly virulent on all canola cultivars currently on the market.
Over the past few years, researchers in Alberta have been trying to better understand the strains of the clubroot pathogen that are able to overcome clubroot resistance. “One of our research efforts is focused on monitoring canola fields across Alberta to understand where resistance is holding up and where it seems to be breaking down, and to characterize the different pathotypes,” explains Stephen Strelkov, professor of plant pathology at the University of Alberta. “We quickly realized that we needed to develop a new classification system for these new pathotypes, as the commonly used pathotype classification systems were unable to identify or distinguish ‘new’ P. brassicae strains that overcome resistance from ‘old’ strains.”
For example, the first of the new resistance-breaking strains of P. brassicae to be identified in Canada was initially classified as pathotype 5 on the existing Williams’ differential set, however this classification did not reflect their virulence on CR canola. Therefore, this new virulent pathotype was called “5X” to distinguish it from the old pathotype 5. However, the term “5X” was soon applied to all strains able to overcome resistance, even after it became clear that not all new strains were alike and some had distinct virulence patterns. This created an urgent need for a new system to identify and distinguish P. brassicae strains and keep up with the emerging new virulence phenotypes that were being identified.
Strelkov and research scientist Sheau-Fang Hwang of Alberta Agriculture and Forestry, developed a new system to distinguish pathotypes using hosts that were more relevant to canola in Western Canada. This new model, called the Canadian Clubroot Differential (CCD) Set consisted of 13 more relevant differential hosts and several canola genotypes, which have proven to be able to differentiate among multiple distinct virulence phenotypes among pathogen populations able to overcome resistance. To make it easier to recognize and distinguish between pathotype strains and virulence patterns, the old number system is combined with a new lettering system. For example, old pathotype 3 is now 3H. The most common of the new pathotypes is 3A, while the name 5X was kept for the first of the virulent pathotypes. “After three years of hard work we now have a Western Canadian foundational model to work from,” Hwang says. “We have been able to divide pathotypes into several groups, which helps breeders better select for resistance. With the foundation in place, we recognize that the CCD model needs refining, but at least now there is a base to move forward from.”
Researchers are also trying to understand how much diversity is in the population, and to determine the relationship and genetic similarities or distinct differences of the pathogen strains. “We are using molecular analysis or genotyping to determine the differences, with pathotype 5X, for example, proving to be genetically very distinctly different from the old pathotype 5,” Strelkov says. “This genetic information is useful, providing us with a bit of a family tree that can help us understand which pathotypes are more closely related or not. This allows us to group the pathotypes and then develop molecular markers than can speed up screening to distinguish between different pathotypes and how they are behaving on resistance. Molecular markers allow us to screen many more samples very quickly, reducing the need for further testing to a subset of plants that are producing strange reactions or to verify our results. Eventually some of this material may be used by diagnostic labs in their testing protocols.”
Another question is whether pathotypes such as 5X or 3A are really “new” pathotypes or not. “We wanted to know if these pathotypes were here from the beginning and went unnoticed, or did they arise through other mechanisms such as mutation or sexual recombination,” Strelkov explains. “We went back to historical clubroot gall collections of eight Alberta counties from 2005 to 2016 to see if these new pathotypes were present, using recently developed molecular markers. In fact, we found that in quite a few of the galls, these new pathotypes were present at very low levels, lurking in the background and favored by planting susceptible varieties to this new pathotype. The results showed that these pathotypes were usually found at very low levels in galls from non-CR canola. They also confirm that the proliferation of virulent strains resulted from selection pressure imposed by planting CR canola, eventually resulting in pathotype shifts.” In 2017 the analysis was expanded to include a limited number of samples from other regions in Saskatchewan and Manitoba. So far the good news is resistance is holding up in those provinces. Additional samples are being collected in 2018 and will be tested again.
Resisting clubroot resistance
Also of note is for the first time in 2017, 20 of 113 fields surveyed in the Peace Region tested positive for clubroot. “We tested samples from those 20 fields using PCR and DNA testing and found that in 15 of 20 fields, the new pathotype 5X was identified,” Hwang explains. “According to the DNA, about 0.005 per cent of pathotype 5X spores were buried in the samples, suggested it is lurking in the soil at low levels, but more intensive rotations increase the selection pressure for these novel pathotypes that can overcome resistance. Applying a new calculation that I just developed for the first time on the number of spores released by a clubroot gall, that translates to over 800,000 pathotype 5X spores from each infected canola plant. We know that each canola plant can have 20 grams of gall tissue, and each gram of gall can contain 800 million spores. This means one mature canola plant could release 16 billion spores.”
Hwang emphasizes that rotations of two or three years between canola are required to help reduce the levels of infections from those 16 billion spores per mature canola plant gall. “Three-year intervals between canola rotations actually result in significantly higher canola yields than a two-year rotation, however, pathogen levels are reduced with both rotations. It’s not just about reducing novel pathogens, it is important to reduce the whole pathogen population in the field because as long as population levels are kept low, then the disease can be delayed and yield losses kept lower. According to a survey we completed in 2015 in four counties in central Alberta on 5,598 fields, most growers are using good practices and rotations, with only about 10 or 15 per cent using very tight rotations of no break or one year in between canola. The results from the 2017 Peace Region survey were similar.”
For growers, there is no single magic bullet, but genetic resistance remains the easiest and most effective clubroot management strategy. However, to maintain the effectiveness of these resources, other good management strategies such as extended rotations of at least two to three years is recommended. “We are researching other strategies such as fumigation to try to reduce population densities, particularly in field hot spots or field entrances,” Hwang adds. “We need to take a holistic view and determine how other factors such as weeds impact clubroot pathogen development. We also know that the pathogen prefers acidic conditions and is less favored in higher pH soils. A graduate student project is underway to trial the potential use of lime amendments and different formulations, with early results very promising. The next major project is to find a way to measure specific density of spores in the field.”
Strelkov and Hwang will continue to screen for new pathotypes and refine the CCD model. A better understanding of the pathotypes helps inform plant breeding and the focus of resistance strategies. “It is important to be breeding for prevalent pathotypes, so the right resistance is there when new canola varieties are released,” Strelkov says. “We continue to do this pathology work so breeders know how to target their efforts and also to have materials with which to screen against. Resistance stewardship and a more integrated approach will be needed for sustainable clubroot management as we try to balance clubroot resistance with evolving pathogen strains and virulence.”